Genetically Modified (GM) Yeast Strains: Unlocking Bound Hop Thiols and Engineering Targeted Fermentation Characteristics

led Image Courtesy of Omega Yeast
S
ettle in this one is dense, but I promise (or at least hope) approachable and engaging!
Often, when I get excited about a topic and can’t wait to experiment with data from the literature and write a post about what I learned (even if it takes me a couple months like this one). In that case, I think there is a good chance many brewers might share my interest in the topic. The GM yeast developments is one of those areas! There’s still so much to learn and explore, but early research and experimentation have convinced me that GM strains can be great tools for brewers to create complexity, new and exciting beers, and new hybrid styles. But GM technology is often approached cautiously, and for me anyway, the best way to form an opinion on something is to learn as much as possible about the topic. I did my best here to do exactly that, looking at GM technology, exploring GM yeast fermentation studies, interviewing the experts engineering GM strains, and reaching out to academic authors for guidance and clarification.
In this post, I explore the topic of genetically modified foods, in brief, to set the stage and create a context to how science is currently being investigated in beer and wine fermentations. I discuss some of the new genetically modified (GM) yeast strains that have been created to pull the curtain back on the how and why questions. I’ll follow up this post with more detailed experiences of these new strains as I have now had the opportunity to brew with them both in small-scale experimental batches as well as scaled-up batches. We even somehow convinced Bissell Brothers to try one of the strains out with us at Sapwood!
Genetically Modified Technology
To educate me for this post, I started first by reading (well, not really, I listened to it on Audible) a book titled Food Fight: GMOs and the Future of the American Diet. I mainly needed a refresher course on what genetic modification was, and this seemed like the only book I could find that was attempting to give both sides of the often-polarizing topic. The book opened by discussing one of the first genetically modified foods approved by the FDA in 1997 called Flavr Savr (much longer ago than I would have guessed). Although flopping, the tomato was engineered to ripen on the vine naturally during transit but still tough enough to resist rotting and mechanical picking. Using science for the crop improvement process was new, and as far as I can tell, it went relatively under the radar. To be fair, though, I was 14 and more concerned with protecting my size 6 Jordan’s from the South Dakota snow than paying attention to the latest agricultural science breakthroughs.
Fast forward to the present day, and the term GMO has become politically charged and polarizing, likely for several reasons like the mistrust of big food businesses, the seemingly unnatural nature of GMOs, and the confusion of information and data out there, to name a few. Nobody comes here to read about politics, I get that, but as I experiment with and learn about GMO yeast strains, I feel like there’s going to be some passionate pushback as well as praise. To approach this subject, I felt like I needed to do a little of my research on its history and place in our society to understand and perhaps form an opinion on gene-altering technology.
Despite such high polarization of GMO foods, the reality is that globally, it is estimated that 70-90% of animals raised to eat are fed GMO foods. The middle of the grocery isles is littered with processed foods made with GMO corn and soybeans (think high fructose corn syrup and soybean oil). GMO foods have already found their way into some of the most sold beer brands out there today, thanks to GMO corn and corn syrup in American light lagers. However, there seems to have been fear or hesitation of introducing GMO foods into beer production initially, whether for marketing purposes or other reasons. The book mentioned above states that Anheuser-Busch in 2008 announced they would not buy any of the $100 million of rice grown in Missouri if GM rice was allowed to be grown anywhere in the state. This was mainly due to the potential cross-contamination of GM plants with non-GM plants used to produce their beer. I wonder if GM yeast strains being engineered today will be looked down upon by some of these more prominent brands already using related science in the grist side of brewing?
When it comes to GM technology finding its way into beer via yeast strains, some of the safety concerns that often are attributed to GMO farming seem distant. For example, a GM yeast strain doesn’t require potentially dangerous pesticides and herbicide spraying like GMO seeds might. These aren’t roundup ready-like GMOs. Instead, researchers essentially remove or add a targeted gene for a specific purpose (Ie. enhance certain esters, remove certain phenols, or enhance biotransformation potential). Although this distinction seems obvious, it also seemed worth pointing out.
Types of GMOs
GMOs are of two varieties, cisgenic GMOs involve only genes from the plant itself or a close relative, and traditional breeding techniques could also transfer these genes. The other type is transgenic, which means the transferred gene usually derives from an alien species that is neither the recipient species nor a close, sexually compatible relative. 1 As for GM yeast strains, some efforts are cisgenic GM strains because the gene pool is not being altered. In other words, a gene is not being transferred that is not naturally occurring in yeast already. We also see a transgenic approach with yeast strains with genes inserted from food-grade bacterial sources or deriving from plants to enhance biotransformation enzyme potential. Not to say one method is better than the other, as both are being utilized in beer and wine fermentations successfully.
Natural Selection Modification vs. Lab Modification
Genetically modified seeds have been used by farmers naturally for years by hand-selecting seeds or hybridizing seeds that performed the best to increase future harvests’ quality and volume. This would be what I view as more of a natural selection modification process, which is still a form of human intervention but in a slower holistic fashion. In a twisted way, altering genes in a lab speeds up this somewhat natural but not natural process. Mutations in genes happen naturally, but not with the precision and speed as labs can now perform with tools like CRISPR (discussed below).
Human intervention into new yeast strains is not new; work has been done to hybridize strains for several reasons like aid in aroma formation (like encouraging ethyl and acetate esters), faster fermentation, uptake of certain sugars, and the ability to ferment in stressful environments. 2 Hybridizing strains are a way to create new yeast strains with specific properties without using more controversial GM technology and likely a process more widely accepted. This is especially true when considering that lager yeast (S. pastorianus) is a natural hybrid between S. cerevisiae and the newly discovered S. eubayanus. 3 However, as I discuss later in the article, there are complexities to the hybridization approach in terms of unwanted or unknown fermentation characteristics that can be avoided with the more targeted GM technology.
Some brewers would prefer this slower and more natural approach to yeast gene modification. In an interview with NPR, Firestone Walker Brewing Co.’s brewmaster (and somebody I look up to) Matt Brynildson commented that we should slow down the direction in which some labs are going related to GM yeast strain engineering.
“The next thing you know, we might be making beer with water, a drum of the cheapest sugar source you can find, and yeast that makes all the flavors that we used to get from barley and hops. That just wouldn’t be fun anymore, If we allow GMO yeast, well, I could think of a hundred more things that I do or don’t want my yeast to do.” – Matt Brynildson, Firestone Walker
I, too, can see the skepticism in producing such expressive yeast strains that you could argue might take some of the romanticism and skill out of brewing. I also agree with this statement regarding what to ask for from these strains; I even suggest one potential avenue of gene modification at the end of this post. But whether we should be doing this is a separate question altogether. At the same time, I also cannot stop thinking about the potential of combining bioconverting enzyme-producing strains with fruit-forward ester-producing strains to create new and potentially explosive flavors and aromas. There’s also a question of where do we draw the line for the use of lab-produced brewing ingredients. For example, exogenous enzymes are already sold and used to alter the natural brewing process (both cisgenic and transgenic), whether for bioconversion (beta-glucosidase enzymes), removing chill haze and gluten (Clarity Ferm), or improving the conversion of starches into sugars (amylase enzyme). All the enzymes mentioned above are lab-produced and perform tasks that the yeast, malt, and brewing process cannot generally do independently. Some, like ALDC (Alpha Acetolactate Decarboxylase), used to limit the production of diacetyl are even produced by a transgenic fungus.
In my experience with test batches of GM yeast strains (which I plan to discuss in more detail in future posts), the result has been more of a way to build in added complexity and less to replace other ingredients altogether. I’m more inclined to be fascinated with new developments and products in brewing research and often try to force-feed them into my brewing experiments. Sometimes I do this with success, changing an ingrained process point, and more often, I don’t see much of an impact at all, adjusting back to the norm. I guess what I am trying to say is that my excitement for GM yeast potential outweighs my reservations.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)
How are these GM strains being created? Gene modification of most yeast strains is being done with a genome editing technology called CRISPR (other methods also exist, as Berkelely Yeast started trials with CRISPR but now utilizes a different set of molecular scissors). The technology took a big step forward in the United States in 2016 when the Department of Agriculture decided it would not regulate a CRISPR-created mushroom. Researchers engineered the white button mushroom to resist browning by targeting a family of genes, which ultimately reduced browning enzymes by 30%.4
CRISPR is now being used across multiple industries for various purposes. What exactly is CRISPR allowing researchers to do? Discussed on a recent Master Brewers Podcast, Charles Denby author of a recent paper titled “Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer,” described CRISPR as a way for scientists to make targeted double-strand breaks in DNA. If there is a part of the genome that is desired to be cut, they can then supply an exogenous piece of DNA, and if that DNA contains similar sequences of the cut ends, then that new DNA can be incorporated into the genome of the new organism.
Bryan Donaldson of Lagunitas noted on the podcast above that there were notes of fruity loops and orange blossoms to the beers made with the modified strains. He went on to say there were good fruity flavors in there without green vegetal flavors, but there was something “fundamentally missing” in the hop flavor. Donaldson suggested that trying to replicate the thousands of hop compounds with two targeted compounds (linalool and geraniol) is not likely to happen. I would agree with this observation.
GM Yeasts in Fermentations
Although this is the first I’ve written about GM strains, and its presence in beer fermentation is relatively new, it’s been around longer than we all probably realize, just not in the alcohol consumption arena. Saccharomyces cerevisiae has an estimated 6,000 genes, and genetically modifying them for specific chemical production and pharmaceuticals studies and production is already being done.5
As it relates to the present-day GM development of yeast strains, each strain’s goal seems to rest in the pursuit of increased biotransformation ability (both with wine and beer ferments) or the suppression of certain unwanted fermentation flavors like unwanted phenolics. However, there seems to be an incredible amount of potential in other avenues of targeted fermentation characteristics.
The wine world has been a little quicker to embrace GM yeast potential (at least in labs). A paper dated back to 2004 successfully engineered a saccharomyces strain to improve L-malic acid degradation. Saccharomyces strains cannot typically degrade -malic acid from grape must. The strain was also found to slightly increase the volatile acidity, ethanol, and glycerol production in test ferments. Sensory tests also found a sensory impact by enhancing the aromas. These “malo-ethanolic” strains were looked at as particularly beneficial of fruity-floral wines and de-acidification of high-acid wines in cool growing climates. 6
Although the wine industry has been looking at GMs for some time, there’s been some resistance from winemakers to use them (as the one tested above didn’t go into production as I understand it). It’s unclear to me how many winemakers are using GM strains currently and they may even be looking at the brewing industry to see how the public and brewers themselves react to the new strains. Especially considering the move in wine circles is going natural, biodynamic, and no intervention. For example with natural wines (like orange wines) no yeast is pitched at all, rather, the natural yeast on the fruit itself ferments the wine (similar in ways to coolship lambic).
In 2007 wine researchers released a paper where the goal was to engineer a wine strain capable of releasing a significant amount of bound wine thiols by overexpressing the bacterial tnaA in S. cerevisiae which creates active beta-lyase activity. Inserting tnaA into VIN13 (wine strain) did result in significantly more bound thiol release than the VIN13 that wasn’t modified.7 Sensory panels also found that the modified strain with higher amounts of now free thiols scored higher in passionfruit, grapefruit, and box hedge flavors. They did note that an increase in the sweaty and cat urine aroma was observed at really high 4MMP thiol levels, suggesting that there could be ideal thresholds for certain desirable flavors with each specific compound.
The authors of the paper state that “Based on our results, we suggest that the cysteine conjugate precursors are transported into the yeast cells, the enzymatic activity takes place inside the cells, and the thiol is released and removed from the cells, either through diffusion or through active transport.” In other words, the precursors from the grapes (or hops) are likely transported into the yeast cells, where the active beta-lyase activity frees the thiols.
To me, when using the results above when thinking about beer and hop ferments, it would make sense to try and get as many the hop-derived thiol precursors into your beer as possible for the start of fermentation. Suppose the activity is happening inside the yeast cell. In that case, it makes sense that adding it late into fermentation when most of the cells have flocculated would result in less of this thiol-releasing bioconversion. This is especially true when considering work done earlier in another wine study that found free thiols were detected after the first day of fermentation and peaked at the early fermentation stages. 8 Meeting consumer expectations through management in vineyard and winery: yeast choice for fermentation offers great potential to adjust the aroma). Possible ways to introduce these bound hop compounds for engineered strains to thrive on could be via pre-boil/boil hopping, whirlpool hopping, and brew day dry-hopping.
Unlocking Bound Hop-Derived Thiol Potential in GM Yeasts
This post (and most of the researchers’ attention) focuses on releasing bound thiols (3MH and 4MMP) and not bound monoterpene alcohols like linalool and geraniol. Although these terpene alcohols have long been great markers in research for determining the hoppiness in beer and play a role in the final flavors of hop-forward beers, increasing these compounds just doesn’t appear to have the same bang for the buck as releasing bound thiols. Thiols are just so fruit-forward in their sensory attributes and, because of their low taste threshold, can have a much more significant impact on the beer than their terpene counterparts. Thiols have also become the hot topic among academic work the past few years, something I welcome!
I’ve written before on potential ways to release bound hop-derived glycosides (like linalool and geraniol), and I still see it as an exciting area of brewing hoppy beers. But the more we learn about thiols, the more I see these terpenes as complexity builders and not the headliner. Using a beta-glycosidase enzyme (to free bound terpenes) like AROMAZYME still seems worth exploring, especially considering that thiols and terpene alcohols can play off each other through synergy, generally enhancing each other’s perception. The other enzyme, beta-lyase, can free bound thiols that might have a more considerable overall effect and that is where GM yeast strains come into play.
What are the two main thiol precursors in hops? The majority of amino acid precursors in bound thiol hop compounds are glutathione-thiol (glut-thiol) and not cysteine-thiol (cys-thiol) conjugates. The majority of glut-thiol precursors in hops is unfortunate because current testing has only been able to free-bound cysteine-thiol conjugates with active beta-lyase activity, the enzyme required to free bound hop thiols (by way of the IRC7 or TnaA gene). The IRC7 gene is likely the best yeast-derived (cisgenic) gene candidate to access and help liberate bound hop-derived cys-thiol precursors. The amount of free thiols, like 3MH, in hops, is often in much higher concentrations than its free form, showing how exciting figuring out how to free them can be.
However, it’s worth noting that Berkeley Yeast has done experiments that freed glut-3MH precursors when overexpressing cysteine–S-conjugate lyases(C-S) lyase activity. Cysteine–S-conjugate lyases are part of a large enzyme carbon-sulfur lyase family, which have been isolated from rats, humans, zebra fish, and Arabidopsis thaliana (flowering plant). 9 I’ll be curious to see if further testing to release glut-thiol precursors is done by targeting and inserting the gene(s) responsible for C-S lyase activity in beer strains, but for now, the focus is on freeing the cys-bound thiol precursors through targeted genes.
A paper dated back to 2013 shows the beta-lyase enzyme inability to break down glutathione conjugates, but rather cysteine conjugates. The paper states, “Therefore, S-glutathione conjugates (also evidenced as precursors of thiols in wine) were not degraded in our experiments.10 In terms of hop variety tested here, the paper found that Cascade exhibited the highest potential of bound 3HA (by releasing cys-3MH). Columbus, Nelson Sauvin, and Saaz extract released less, but still 29, 23, and 126-fold higher than without the beta-lyase enzyme.
Interestingly, the study’s beta-lyase also released the skunky MBT thiol at high levels (0.584 ppm for Nelson and 0.454 ppm for Columbus), both well above the extremely low MBT threshold 0.0000044 to 0.000035 ppm. 11
The MBT results above show that the hop variety we are using with strains engineered to have the IRC7 or TnaA gene and subsequent beta-lyase activity can impact the beer positively or negatively. Ideally, it seems, we would want hops with high concentrations of bound cys-3MH, which could be freed to 3MH while also avoiding hops that could produce high amounts of MBT (like Nelson). It’s interesting to note that some commonly used strains in hop-forward beer styles have been found to have inactive IRC7. To be clear, traditional brewing strains can still have the IRC7 gene, it just might be a mutated version that doesn’t have any activity, showing where GM technology can come into play (by switching it on).
The focus isn’t entirely on the IRC7 gene; Other organisms also contain genes that code for beta-lyase enzymes. In an early demonstration of how transgenetic material can be used to increase yeast beta-lyase activity, researchers at the Australian Wine Research Institute inserted a gene from E. coli into a wine yeast’s genome and showed that it had a lot more beta-lyase activity than its parent strain. For example, in the paper above that tested Cascade, Nelson, and Saaz, the beta-lyase enzyme used was derived from apotryptophanase (TnaA) from Escherichia coli. Using a gene from Escherichia coli would be an example of a transgenic modified approach as the gene responsible for thiol biotransformation via beta-lyase activity is coming from a non-yeast source. In contrast, when changing a yeast strain by adding and overexpressing the IRC7 gene, this is a cisgenic form of modification because the IRC7 gene already exists in yeast strains (likely more wine strains).
In terms of wine fermentations, this particular gene (TnaA) was also found to free bound cys-4MMP precursors suggesting it might be an excellent gene for releasing both 3MH and 4MMP.12 Berkeley Yeast has taken a transgenic approach to engineering brewer’s yeast for extra beta-lyase activity. Tropics™, which is a strain discussed in the post, contains a food-grade beta-lyase. On the other hand, Omega Yeast with its thiol-producing strain is using the IRC7 approach (derives from yeast already), also discussed in the post.
What is the threshold of these thiols, and what are the concentrations in hazy, hoppy beers? In 2021 researchers found that after comparing six hazy commercial IPAs for hop-derived compounds compared to West Coast IPAs, found a significant increase with the hazy beers.13
Interestingly, however, the thiol concentrations in these heavily dry-hopped beers were below the threshold in both 3MH and 3MHA. The thiol 4MMP was found above threshold across the average of hazy IPAs tested, showing that there is likely enough free 4MMP in most hazy hop varieties (Cheater Hops as I like to call them) to get above the threshold to have an impact on flavor and aroma. However, when it comes to 3MH active beta-lyase activity is probably required to get you above the threshold to free bound cys-3MH thiols unless you are pounding the beer with dry-hops. This is where GM yeast strains can play a significant role in releasing bound 3MH, potentially setting up even more 3MH conversion. A side note from this study cited above is that the thiol concentrations were not impacted much after being centrifuged, suggesting they are much more polar than some of the greener and woody hydrocarbons from hops (like myrcene which was significantly reduced after centrifugation).
| Thiols | Aroma/Flavor | Threshold (ppb) | Average in Commercial Hazy IPAs Tested (ppb) |
| 4MMP | Grapefruit | 0.0015 | 0.15 |
| 3MH | Box Tree, Black Currant | 0.055 | 0.03 |
| 3MHA | Passionfruit | 0.005 | 0 |
It makes sense to take a step back then and summarize what’s going on here with these modified strains. What is being added, taken away, and overexpressed? When it comes to biotransformation potential in the cisgenic approach, labs are adding the IRC7 gene and (in some cases) overexpressing it, meaning it’s there but with elevated levels of enzyme activity. Inserting this gene into a strain like a London Ale III equivalent not only adds the ability to release bound hop thiols in a strain that otherwise doesn’t have the potential, but amplifies. It also keeps the other desirable traits of a strain like London Ale III in place (mouthfeel, haze, attenuation, etc.).
The same is true in the transgenic approach. Still, a lab is likely taking another source of active beta-lyase enzyme activity utilizing a protein deriving from another source like another microbe or plant. By inserting this non-yeast derived protein into a beer yeast strain (and overexpressing it), you are (like with IRC7) setting up the potential of releasing bound hop thiols. It seems that the transgenic approach may be a more efficient approach to act on the bound hop substrates, freeing a more significant amount. For example, in Berkeley Yeast’s trials with Tropics™, they measured more than a 100-fold increase in released otherwise bound 3MH than a traditional highly active strain IRC7 gene strain. However, new strains are still being developed and tested with the IRC7 gene. I can tell you through my own trials that the Tropics™ strain is not only performing on paper, but its sensory impact was obvious too, in a very good way.
How are labs like Berkeley’s finding other potential genes to experiment with that might have enzyme activity capable of bioconverting? There are programs available to researchers like BLAST (genomic alignment tool) to insert gene sequences and look for similar ones. For example, put in a beta-lyase sequence and look for similar amino acid sequences from different sources that encode other beta-lyases. This can help target researchers’ approach to what kinds of active genes could force the beta-lyase enzyme activity in ferments. Two enzymes with similar amino acid composition may have wildly different activities. Some will be active when produced in yeast and others will be inactive. For example, in the Berkeley strain case, they used this approach to find other active bioconversion genes. They had to try many different beta-lyases before finding one that was especially good at releasing thiols from
both gluthathionylated and cysteinylated substrates, which is even more impressive considering previous research was just freeing only the cys-substrates.
GM Test Batch w/ Cascade/Columbus Whirlpool fermented with Berkeley’s Tropics was screaming tropical fruit pre dry-hop!
Potential for Thiol Precursors in Hops
In an email conversation with Aurelie Roland, who authored an excellent paper testing the different bound concentrations of both cys and glu-thiol precursors (results in the chart below), I asked if the heat from the kettle would be enough to help cleave some of the bound thiols. I’ve theorized before that maybe whirlpool temperatures might help to release some of these compounds, but that doesn’t seem to be the case. Roland explained that heating is not sufficient to break the chemical bond between cys and glu-thiol precursors. In other words, releasing bound thiols is solely a job for an enzyme. Other methods for accessing these bound thiols could occur with beta-lyase enzyme additions (not via yeast) similar to the addition of a glucosidase with AROMAZYME. However, I can say when using products like Scott Lab’s Expression Aroma, which is described to extract “aroma precursors in white grapes such as thiols,” has not had even close to the sensory impact as something like the Berkeley’s Tropics™ GM strain. It’s unclear to me what exact enzyme Expression Aroma is, perhaps it’s not beta-lyase?
I like Roland and his team’s work so much because the results show how much thiol potential is in hops that are not being utilized in traditional ale ferments due to lack of beta-lyase activity. Below is a chart of the hop varieties they tested, comparing the free and bound concentrations of 3MH and 3MHA compared to the bound cys-precursors. I didn’t include the amounts of bound glu-3MH and glu-4MMP because we learned earlier in this post the inability of the beta-lyase enzyme to break down glutathione conjugates. Although, you can see why Berkeley Yeast might be onto something in their trials freeing both bound categories. Why? Because Saaz led the pack with an incredible 20,678 ppb of bound glu-3MH! 14
Not only do the charts above help us to see the hidden potential in specific hops, but it makes it clear what hops are better for the hot side and which ones for dry-hopping. Hops high in free 4MMP would be great dry-hop varieties like Citra®, Eureka™, and Simcoe®, and hops like Calypso™ and Saaz are probably great whirlpool hops when used in conjunction with active beta-lyase activity. The paper also shows why so much of the focus is on freeing bound 3MH because of the precursors’ dominance compared to 4MMP.
Potential in Malt for 3MH Precursors
Surprisingly, hops aren’t the only brewing ingredient that has bound fruity 3MH thiol potential. In a paper studying thiols, it was found that 3MH was being detected in unhopped beer but not in unhopped wort, suggesting that malt-derived 3MH precursors are coming from the malt. 15 But how do we know what types of malts?
A study was done in France where the authors investigated 3MH and 4MMP thiol precursors in six-row malted and non-malted barley samples. They found that G3MH (a 3MH precursor) was higher in malted than non-malted barley, suggesting an in-situ formation during malting. This may be important because it indicates that malted grains over unmalted are beneficial when encouraging GM strains’ bioflavoring process. Compared to large percentages of unmalted grains, 3MH increases in the boil, with thiols from malt playing a more significant role than we would think. The current study hypothesizes that less than 3% of thiol precursors in malts could lead to more than 64% of 3MH in the final beer because of the evolution during brewing. 16 I imagine this study done with a GM yeast strain with either an overexpressed IRC7 or other bete-lyase gene would see these numbers inflate even more.
It would be fun to see more research into what factors might be playing a role in 3MH precursors in malted grains. One theory is that lowered kiln barley might have more 3MH precursors than barley kilned at warmer temperatures (pale malts having more than crystal malts, for example).
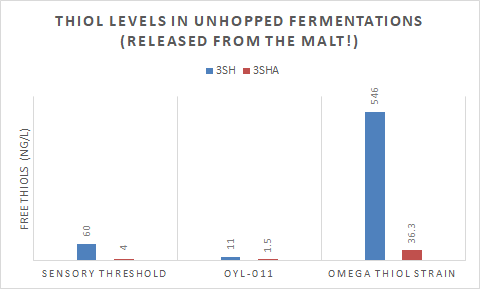
While it’s unclear to me now the exact concentration of 3MH precursors in different malt types, the data supplied by Omega Yeast’s strain called Cosmic Punch™ (chart below) shows how efficient a GM strain engineered to free bound 3MH can be. Their to-be-named thiol producing strain (IRC7 overexpressed) fermented in unhopped wort freed an enormous amount of 3MH from just grain precursors, well over threshold for both 3MH and 3MHA. The chart shows how little freeing their British ale strain is capable of, likely the same for many other traditionally used strains for hoppy beers.
What if you pair the results from Omega with the hops high in bound 3MH, would you get a beer with thiol concentrations that are much too high to be enjoyable? The answer is I don’t know and I guess it’s up to us to find out! I can say in test batches with Berkeley’s thiol driving GM strain which is also freeing an enormous amount of 3MH from malts, did produce an extremely aromatic beer with lots of tropical fruit (guava and passionfruit). I also paired the beer with hops high in bound 3MH, which likely drove the thiol count even higher. I will say, to some, the beer was a little white peppery or spicy as well as super tropical.
I think there is a lot of experimentation opportunities for brewers here to figure out first, the profiles these strains can produce, and second, try to appropriately place them in recipes for building balanced and but highly aromatic beers. Maybe copitching them to build in a little more balance as possible example.
Side Enzyme Activity and Other Observations
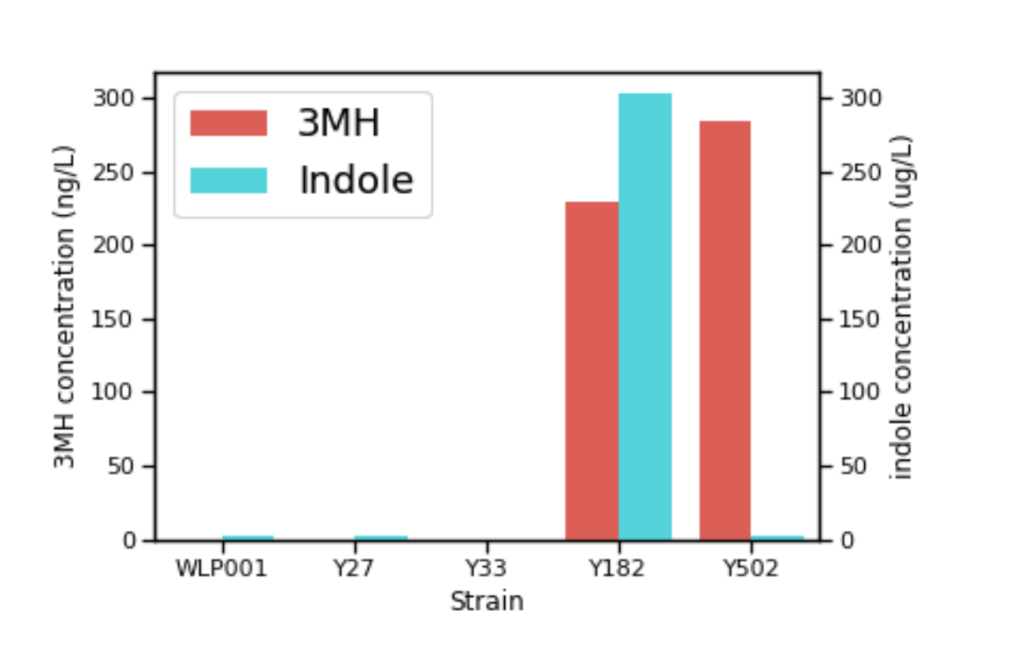
How might adding or altering one of the approximately 6,000 genes in a yeast strain impact the other genes and how they operate? It’s not as simple as just inserting a gene and getting the outcome (and only the outcome) you desire. Enzymes are specific in their respective activity, so when you overexpress something, you’ll likely get more of everything that gene is doing (both good and bad). A great example of this is when Berkeley Yeast was going through trials on creating a thiol-enhancing strain called Tropics™. They initially repeated the work done by the Australian Wine Research Group and they found that TnaA beta-lyase activity can release bound thiols and convert tryptophan (an amino acid) to indole. This is something they already knew; however, they just didn’t anticipate the resulting overexpressed strain would release as much indole as it did.
What’s indole? Simply put, it smells like poop at high concentrations. Indole is likely present in both beer and wine fermentation at low concentrations. At these lower levels, indole can add to the complexity. In early Berkeley experimental fermentations with the beta strain using the wine group’s TnaA they were getting great tropical notes but also getting fecal notes over the threshold (as the chart below shows). After they figured out how to get rid of the indole, they eventually found a different food-grade beta-lyase, which is what they used to make Tropics™. I can attest after a few small test batches that the indole is gone and the strain is incredibly fruity.
Omega Yeast is nearing the release of their strain of a traditional hazy culture with the IRC7 gene both inserted and overexpressed to push the cisgenic biotransformational release of bound hop thiols above sensory levels. An obstacle for Omega was not in the form of indole but with elevated levels of sulfur compounds being produced, which can also approach sensory threshold in lagers and wheat ales. Laura Burns, Director of Research and Development at Omega Yeast, told me that “we are finding that the final version of this strain has elevated sulfur production, though it is cleaning up at the end of fermentation. We are working on targeting the right amount of IRC7 activity to enhance only thiols and make sure that these sulfur compounds don’t approach sensory levels.” I am excited to give the completed strain a run in the future!
Genetically Modified (GM) Strains vs. Traditional Hybridization
Just because a strain was created in a lab does not make it a GM strain; as mentioned earlier in the article, strains can also be mated together through hybridization. Yeast creation through hybridization well predates CRISPR creations but comes with its own set of challenges.
Why not just use this more traditional approach vs. the new GM engineering approach? Simply put, it’s not as targeted. Think of yourself as a product of both of your parents, you embody traits of both of them (probably some good and bad!), yeast strains are no different. For example, if you create a hybrid strain with a bioconverting wine strain and a hazy IPA strain, you’ll likely end up with a new strain with the IRC7 gene making it capable of releasing bound flavors. The new strain might also take on other aspects of wine strain like phenolic production, flocculation traits, alcohol tolerance, etc.
A great example of this is a hybrid strain Omega Yeast sent to Sapwood to test out mating a hazy IPA strain and a bioconverting wine strain called Maxithiol. The Maxithiol wine strain “has the ability to produce aromatic thiols which contribute to significant fruity esters of tropical fruit and passion fruit to the finished wine.” The idea, you get the same hazy, hoppy characteristics from the ale strain and the thiol-releasing capabilities from the wine yeast. A bonus, because it’s a hybrid strain and not simply a copitch, the difference is the hybrid will get you the same results in the consecutive batch it’s harvested and pitched into. In contrast, a copitch would likely see drift and dominance from one of the two strains used in the blend, ultimately changing fermentation characteristics in future batches.
How did this hazy/wine yeast hybrid perform? We noticed a few behaviors with the strain that we don’t see in traditional ale strains used for making hazy IPAs. The hybrid strain was more attenuative than what we were used to with the source strain, likely resulting from a more aggressive fermentation capability in the Maxithiol strain. This could be offset with processes or ingredients (mash temperatures or maltodextrin, for example).
We noticed in test batches of the hybrid strain that we couldn’t harvest the yeast in volumes we were accustomed to in normal ale fermentations. Usually, we can harvest yeast from a 10-barrel batch and have enough yeast for a couple of 20-barrel ferments of IPA and DIPA. With the hazy/wine hybrid strain, we found a remarkably high yeast health percentage (90%+). However, the yeast volume we got was nearly half of what we usually get with a traditional non-GM London Ale III strain, making it tricky to re-pitch into future batches as planned. However, I will say our referments with the yeast performed as expected. Again, it appears that Maxithiol is not as flocculant as our usual strain, another trait sneaking its way into the hybrid strain. Yeast taking longer to flocculate out like this in heavily dry-hopped beers can, in our experience, enhance the vegetal bitterness bite and distract from the juicy smooth NEIPA flavor we are after.
Our yeast cell counts (thanks to Michael Tonsmeire’s microscopic abilities) with our usual RVA Manchester Ale strain and the Omega’s hybrid strain varied greatly. You can see an incredibly high yeast viability rate in the hybrid strain, but the total pounds harvested is significantly less. More time cold could have perhaps sped up the harvest poundage (maybe at the expense of some yeast viability), but time is valuable in commercial batches.
| Yeast | Harvest Amount (lbs.) | Cells (billion/mL) | Viability |
| Omega Hazy/wine Hybrid | 75 | ~.285 | 98% |
| RVA Manchester | 90 | ~.900 | 76% |
| RVA Manchester | 110 | ~0.750 | 80% |
| RVA Manchester | 100 | ~0.825 | 78% |
The last thing I’ll say about (and I don’t even know if this matters at all) is that the hazy/wine hybrid strain was also very green at the time of harvest (picture below). It almost looked like it had hops in suspension, similar to doing a drop of the cone with yeast and hops present. This is more just an observation than anything.
I bring up our results with the hybridized strain because it shows clear advantages to the CRISPR GM engineering approach, which is much more targeted in its fermentation outcomes. The GM approach to creating new strains allows control (after understanding each gene’s role) of specific traits from other yeast strains or bacteria. For example, rather than creating a hybrid strain with Maxithiol and a hazy strain to get the IRC7 gene, the GM approach allows you only to insert this gene into the hazy strain, getting you beta-lyase activity without the other outcomes we experienced in the hybrid. This precision is enhanced even more when the gene itself inserted (or replaced) performs only one job. In other words, if inserting a gene that only produces a single ester and nothing else, you know that the new GM strain won’t have any other side activities. This sounds easier than it is, but it’s a helpful way for me to understand and appreciate the difference between the two different approaches to creating new yeast strains.
Traditional Strains with Beta-Lyase Potential
Does it have to be a GM yeast to be able to unlock bound hop thiols? A poster was released at the 2020 World Brewing Congress in a joint research project with white Labs and Oregon State University, looking at what potential yeast strains may have the IRC7 gene naturally with the specific mutations that may release bound hop thiols. They found the strains BE057 (third highest), BE062 (second highest), and BE088 (highest) did surprisingly have the gene and ability to bioconvert. The three strains were found to have significantly greater activity than a wine strain called MaxiThiol, which is selected because of its ability to release bound grape-derived hop thiols.
However, leaning on experimentation and discussion with Nick Harris from Berkeley Yeast, that even highly active IRC7 variants containing mutations in their trials were not producing beers with thiols at sensory levels. This is not to say that strains with higher levels of IRC7 activity cannot make distinguishable beers between a strain without the gene inserted. Using a strain capable of releasing any bound hop compounds should only help build complexity into a beer by increasing thiols into ranges close to or over the threshold. Even under the threshold, the now freed thiols may boost overall flavor through synergy with other extracted hop components. Again, I’m looking forward to testing Omega’s new strain in this overexpressed IRC7 area!
Although I’m excited to see where more of this experimentation goes, I’d love to see traditional strains with active IRC7 genes tested in beer solutions with hops with a higher percentage of bound thiols to see their potential played out.
ATF1 and the 3MH to 3MHA Conversion
Because this post hasn’t already been technical enough, there’s one more area I’d like to cover because it’s one I’ve been interested in for some time in seeing the benefit of increasing the hop-derived thiol 3MH for the potential for the passionfruit-like 3MHA thiol. The problem, what facilitates this conversion once we have 3MH present? Again, we can look at older wine studies to get a better sense of what is required to get this tropical conversion to take place. In 2006, wine researchers tested and confirmed that the ATF1 gene and subsequent enzyme activity are responsible for this conversion (particularly Atflp enzyme). In the study, VIN13 was used as a control and a strain constructed with ATF1 gene inserted and overexpressed. Although VIN13 converted 3MH to 3MHA on its own (~100 ppb in the solution), the strain with ATF1 was a monster producing nearly 700 ppb. 17
So this is great, right? We seem to have now the ability to use hops high in bound 3MH; we can ferment with GM strain designed to release the 3MH at high rates and incorporate an overexpressed ATF1 gene to convert the 3MH into 3MHA for passionfruit bombs, right? Not exactly; Berkeley Yeast did exactly that and found that ATF1 has a broad substrate range, so it acts on many different alcohols. So while it may acetylate 3MH to 3MHA, it also made a lot of isoamyl acetate and ethyl acetate when overexpressed. This again shows that although the enzyme activity is particular when amplifying an activity, it also boosts all the enzyme-related activity.
Not all is lost in the quest for yeast-driven passionfruit thiol creation, as Berkeley Yeast was awarded a grant from the USDA to identify acyltransferases that specifically convert 3MH to 3MHA. This is being done in collaboration with Thomas Shellhammer at OSU.
Omega Yeast’s POF- Project
Biotransformation via active beta-lyase activity-freeing bound hop compounds isn’t the only role for GM yeast strains. The goal is to insert and perhaps overexpress the gene responsible (like IRC) for bet-lyase in bioconversion strains. You can also replace genes inactive in other strains with active ones in the parent strain, ultimately changing the strain’s fermentation profile. Omega Yeast has been exploring this area by essentially turning off the gene responsible for creating spicy and clove-like phenols common in some Belgian and Hefeweizen strains (the FDC1 gene).
Replacing an active gene with an active one responsible for phenol production allows the other fermentation characteristics to shine, like fruity ester production that might otherwise be buried under all of the spicy phenolics. This would be another example of a cisgenic GM yeast because the gene replacement is being done by ones already found in yeasts (strains with inactive FDC1 gene replacing an active FDC1 gene).
Targeting and replacing an active FDC1 gene is something I welcome coming from somebody who has done many experiments copitching wine strains known for biotransformation ability with traditional ale strains. Although these test beers could have unlocked bound compounds from the hops in small concentrations due to enzyme activity coming from the wine yeast, any possible fruity result was hidden behind competing phenol production from the wine strain.
Specifically, Omega has released two strains to the public that use this light switch approach to turning off the phenolic FDC1 gene in a Belgian strain and a Hefeweizen strain. I’ve been lucky enough to brew with both of these strains on both the experimental and commercial batches. Although I’ll likely follow up this post with much more recipe/results from the strains mentioned in this post, I can say that overall, these two strains in our test are subtle complexity builders and not creating fermentation characteristics that completely dominate the beer. In other words, another tool in the brewing toolbox to help achieve whatever it is the end goal is.
We used the Sundew strain in a Sapwood/Bissell Brothers collaboration, the results of which are discussed in the Graining In a podcast hosted by Noah Bissell and Matt Robinson.
Below are the two strains now available from Omega Yeast with their description.
Bananza Ale (OYL-400) – Bananza’s ripe banana flavor boosts tropical character in beer. Bananza is great for dropping more banana notes into your pastry stouts, milkshake IPAs, fruited sours and other modern tropical fruit-driven styles.
Sundew Ale (OYL-401) – Sundew has luscious strawberry, passion fruit and stone fruit esters, which combine to support desirable notes in modern fruity hops. Sundew sets a great foundation for hoppy styles like juicy pale ales, west coast IPAs or hazy IPAs, or its jammy profile can be paired with more malt-forward stouts, mild,s and brown ales. Think versatile like West Coast Ale I (OYL-004), but jammier.
Commercial Yeast Strains Available
Because I’ve briefly mentioned different GM strains (and one hybrid strain) that have been engineered at times throughout this post, below is the list of strains that were the focus of this article and used in test batches at the brewery. The tasting notes are broad as I plan to follow up this post with detailed thoughts on the strains.
If you want to try out these strains, stay tuned for an announcement from Omega releasing their hazy/wine hybrid strain. Berkeley Yeast has informed me that they are selling Tropics™ if you reach out to them directly. If you give these a try, leave a comment telling everyone how you used the GM strain and what your results were!
|
Yeast Name |
Gene |
Targeted Enzyme Activity |
Class of Yeast |
Purpose |
Notes |
|
Omega Hazy/Wine Hybrid |
IRC7 (Maxithiol parent strain) |
beta-lyase |
Traditional Hybrid |
Free bound hop and malt cysteine-thiols |
Creates complexity rather than dominating thiol releasing. |
|
Omega IRC7 + Cosmic Punch™ |
Overexpressed IRC7 |
Enhanced beta-lyase |
Cicgenic GM |
Free bound hop and malt cysteine-thiols |
Omega Yeast reports 3MH levels x100 typical levels. |
|
Overexpressed C-S lyase |
Enhanced beta-lyase |
Transgenic GM |
Free bound hop and malt cysteine-thiols |
Berkeley reports 3MH levels x100 typical levels. |
|
|
FDC1 Swap |
FDC1 Turned Off |
Cisgenic GM |
Remove phenolic producing capabilities |
Allows red-fruit like esters to shine, but subtle. |
|
|
FDC1 Swap |
FDC1 Turned Off |
Cisgenic GM |
Remove phenolic producing capabilities |
Allows banana-like esters to shine, but subtle. |
Other Potential Avenues for Gene Modification
What other areas could gene modification with yeast strains be used in the production of beer? I don’t have a lot of great pitches (see what I did there?), but one area that came to mind when reading through some of the wine research is the potential for increased glycerol production from a GM strain. Although the increase in glycerol production (which can increase the viscosity and mouthfeel) in the 2004 wine study mentioned at the start of this article was slight (1 g/L), it does raise the possibility for high glycerol producing strains. Perhaps a yeast strain capable of producing considerable amounts of glycerol could increase the mouthfeel and sweetness perception enough in hazy IPAs to offset a lower final gravity, reduce caloric intake, and improve the overall drinkability. In my experience, hoppy beers finishing with high final gravities (1.025+) can taste great (in fact, I prefer them), but consuming much of them can be palate taxing. Getting the body and mouthfeel from glycerol while at the same time reducing the final gravity is something I’d like to experiment with.
However, it’s likely not that easy as modifying a strain to overexpress the glycerol-producing gene was tested successfully in wine, but it came with unintended consequences. The study found that test ferments created too much acetaldehyde, acetoin, and acetic acid as a side-effect of the overexpression. 18 . Showing how difficult it is to create these strains!
I’ve also been interested in cold ferments and the impact the reduced temperatures can have on thiol retention to make highly flavorful and aromatic IPLs. Test batches with a strain called Tum 35 with minimal sulfur production (which can compete with hop aromatics) produced a great IPL at the brewery in my opinion (although it seems customers don’t love the term IPL). This particular beer also is by far the beer that’s held onto its fresh hop character better than anything I’ve brewed before.
The science that inspired the test batches in this area found that brewed test beers with Mosaic hops at the onset of fermentation and measured thiols concentrations for 3MH and 3MHA. The authors found that almost twice the amount of 3MHA was calculated in the beer fermented at 59°F (15°C) compared to the one at 71°F (21°C) with a wheat beer yeast (Tum 68). Specifically, the tropical and desirable 3MHA (converted from 3MH) went from 4 ng/l to 8 ng/l at the lower temperature. This might not seem like much, but with a low threshold for 3MHA (9 ng/l), you can see why it’s important. 19
I’m not sure if the greater amount of conversion from 3MH to 3MHA is due to the beer’s reduced temperature encouraging compounds to stay in solution or if the lower temperature is somehow impacting the enzyme active ATF1 gene. Either way, it would be fun to take lager yeasts like Tum 35 and insert an active IRC7 gene to see if the colder ferments help increase bound 3MH concentrations and potentially maximize conversion to the passionfruit 3MHA. Maybe we could make IPLs cool again if this is the case and not have to call them Cold IPAs to sell them! I’ll be testing out a GM lager IRC7+ strain from Omega to potentially get an amped up version of Thiol Driver!
Areas for Experimentation
Copitching strains like Berkeley Tropics™ strain or the soon-to-be-released overexpressed IRC7 strain from Omega with a known ATF1 high producer like Vin13 to encourage greater concentrations of 3MH and subsequent 3MHA levels than what a typical strain might be able to achieve. Better yet, inserting the ATF1 gene from Vin13 (not overexpressing it due to the previous tests in this area) and the over-expressed IRC7 gene into a hazy strain to accomplish this without the phenolics that could occur with the Vin13 strain.
Wine Hybrids is an obvious area of experimentation here to converge the science of earlier years of wine GM yeast production and more recent beer GM yeast science. Creating beers with heavy whirlpool additions of high bound 3MH varieties paired with active fermentation additions of either wine juice alone or juice and skins to increase the precourours even more while also adding another layer of wine-like flavors. New products like Phantasm Powder (pulverized freeze-dried wine grapes) could also be considered for even more thiol-enriching potential. Rather than dry-hop these beers, you could add wine juice early into fermentation to play up this hybrid beer even more and let the yeast do the rest! I may or may not already be trying this. Could Grape Ale be a thing? With your help, we can do this! Maybe age these in wine barrels with your preferred mix-fermentation strain for a year or two and see what happens?
Thanks!
I want to say thank you to both Omega and Berkeley yeast for sending samples of their hybrid and GM strains. I want to specifically thank Laura Burns (Omega) and Nick Harris (Berkeley) for answering question after question through emails and various phone conversations. It took some time to put their complicated and impressive work into a language I could understand and write about with a hint of confidence!
Cheers!
Scott
Key Findings
- There are two types of GM yeast created using CRISPR technology. Cisgenic GMOs involve only genes from the plant itself or a close relative, and traditional breeding techniques could also transfer these genes. The other type is transgenic, which means the transferred gene usually derives from an alien species that is neither the recipient species nor a close, sexually compatible relative. Both are being utilized in creating thiol-releasing beer strains.
- The wine industry has been researching and creating strains dated back to 2004, nearly 20 years before the beer industry.
- The enzyme beta-lyase is required to free bound hop thiols, specifically, the cyc-precursors (cys-3MH and cyc-4MMP). Cys-3MH precursors are found in a dramatically higher concentration than 3MH.
- The hop-derived thiol 4MMP is available in hops in its free state in much higher concentrations than 3MH. Thus, using hops rich in 4MMP (like Citra®, Eureka™, and Simcoe®) are better choices for dry-hopping where enzyme activity is not required to free bound thiols.
- Hops (like Calypso™ and Saaz) have a significant concentration of bound cys-3MH and are likely better suited for kettle and brew day dry-hop additions. Especially considering the enzymatic activity responsible for releasing bound compounds is taking place inside the yeast cell, likely requiring active fermentation to free the bound thiols (with GM or hybrid strains).
- The primary two genes used to create GM strains for biotransformation purposes are IRC7 and TnaA. Both allow for beta-lyase enzyme activity during fermentation to free bound thiols.
- Genes like IRC7 and TnaA can not only be inserted at elevated levels to enhance the biotransformation potential. However, this will also amplify the other traits the particular gene performs.
- Malt also has 3MH precursors, which can be unlocked with GM strains. Lowered kilned barley likely has more of these precursors (pale malts).
- The thiol 3MH can be converted to 3MHA (which has a passionfruit flavor), but this conversion requires the ATF1 gene, which is likely present in most strains but at different expression levels (the wine strain VIN13 converted more 3MH to 3MHA, for example).
- Unlocking bound hop-thiols is not the only use for GM yeast strains. For example, Omega Yeast’s POF- Project replaces the active FDC1 gene responsible for phenolic flavors with an inactive version of FDC1. This turning off of FDC1 prevents phenolic flavors from occurring during fermentation.
Footnotes
- Schouten, H. J., Krens, F. A., & Jacobsen, E. (2006). Cisgenic plants are similar to traditionally bred plants: international regulations for genetically modified organisms should be altered to exempt cisgenesis. EMBO reports, 7(8), 750–753. https://doi.org/10.1038/sj.embor.7400769
- Gibson, B., Geertman, J. A., Hittinger, C. T., Krogerus, K., Libkind, D., Louis, E. J., Magalhães, F., & Sampaio, J. P. (2017). New yeasts-new brews: modern approaches to brewing yeast design and development. FEMS yeast research, 17(4), 10.1093/femsyr/fox038. https://doi.org/10.1093/femsyr/fox038
- Libkind D, Hittinger C, Valerio E, Goncalves C, Dover J, Johnston M, Goncalves P, Sampaio J (2011) Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci USA 108:14539–14544
- Waltz E. Gene-edited CRISPR mushroom escapes US regulation. Nature. 2016; 532(7599):293–293. https://doi.org/10.1038/nature.2016.19754 PMID: 27111611
- Borodina I, Nielsen J. 2014. Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol J 9: 609-620
- Volschenk, H., Viljoen-Bloom, M., van Staden, J., Husnik, J., & van Vuuren, H. (2017). Genetic Engineering of an Industrial Strain of Saccharomyces cerevisiae for L-Malic Acid Degradation via an Efficient Malo-Ethanolic Pathway. South African Journal of Enology and Viticulture, 25(2), 63-73. doi:https://doi.org/10.21548/25-2-2183
- Swiegers, J. H., Capone, D. L., Pardon, K. H., Elsey, G. M., Sefton, M. A., Francis, I. L., & Pretorius, I. S. (2007). Engineering volatile thiol release in saccharomyces cerevisiae for improved wine aroma. Yeast, 24(7), 561-574. doi:10.1002/yea.1493
- Swiegers JH, Francis IL, Herderich MJ, Pretorius IS. 2006a.
- Lash, L. H., R. M. Nelson, R. A. Van Dyke, and M. W. Anders. 1990. Purification and characterization of human kidney cytosolic cysteine conjugate beta-lyase activity. Drug Metab. Dispos. 18:50–54.9
- Gros, J., Tran, T. T., Collin, S. (2013). Enzymatic release of odourant polyfunctional thiols from cysteine conjugates in hop. Journal of the Institute of Brewing, 119(4), 221-227. doi:10.1002/jib.80
- Irwin, A. J., Bordeleau, L., and Barker, R. L. Model studies and flavor threshold determination of 3-methyl-2-butene-l-thiol in beer. J. Am. Soc. Brew. Chem. 51:1-3, 1993.
- Tominaga T, Masneuf I, Dubourdieu D. 1995. A S-cysteine conjugate, precursor of aroma of white sauvignon. J Int Sci Vigne Vin 29: 227–232.
- Biendl, M., Schmidt, C., Paul Maye, J., & Smith, R. (2021). New England IPA – the hop aroma champion of beers. MBAA Technical Quarterly, 58(1). doi:10.1094/tq-58-1-0308-01
- Roland, A., Delpech, S., & Dagan, L. (2017). A Powerful Analytical Indicator to Drive Varietal Thiols Release in Beers: BrewingScience, 70, 170-175
- Kishimoto, T., Morimoto, M., Kobaysashi, M., Yako, N., & Wanikawa, A. (n.d.). Behaviors of 3-Mercaptohexan-1-ol and 3-Mercaptohexyl Acetate During Brewing Processes. American Society of Brewing Chemists, 192-196.
- Roland, A., Delpech, S., Reillon, F., Viel, C., & Dagan, L. (n.d.). First Identification of Cysteinylated and Glutathionylated Precursors of 3-Mercaptohexan-1-ol in Barley. EBC 2017 Presentation
- Swiegers, J. H., Willmott, R., Hill-Ling, A., Capone, D. L., Pardon, K. H., Elsey, G. M., . . . Pretorius, I. S. (2006). Modulation of volatile thiol and ester aromas by modified wine yeast. Flavour Science; Recent Advances and Trends, 113-116. doi:10.1016/s0167-4501(06)80027-0
- Goold, H. D., Kroukamp, H., Williams, T. C., Paulsen, I. T., Varela, C., & Pretorius, I. S. (2017). balancing act between ethanol and glycerol production in low-alcohol wines. Microbial biotechnology, 10(2), 264–278. https://doi.org/10.1111/1751-7915.12488
- Haslbeck, K., Bub, S., Schönberger, C., Zarnkow, M., Jacob, F., & Coelhan, M. (2017). On the Fate of B-Myrcene during Fermentation – The Role of Stripping and Uptake of Hop Oil Compounds by Brewers Yeast in Dry-Hopped Wort and Beer. BrewingScience, 70, 159-169.



-610x915.jpg)
Personally- I’m VERY excited for this next frontier with yeast… and your article and the description of the IRC7 research has only fueled that excitement.
I don’t share Matt Brynildson’s (or anyone else’s) reservations about it all- to me, it is just another tool in the arsenal for anyone trying to make something new, different, and/or exciting. If you want to keep making beer the traditional way- that’s great, but please just don’t be a luddite about it. CRISPR and other forms of gene editing are just a tool. They are not inherently good/bad. They just need to be applied properly. People that reject it on principle are generally just under-informed on the topic and buying into fear-based marketing from the activist “non-gmo project” and Organic Consumers Associations. I had some email correspondence with Imperial Yeast a while back about why they are “organic” and it was just a bunch of marketing garbage- they had no evidence of their product being superior in any way because of its “organic-ness”. Never bought anything from them as a result. On the other hand- I’m happy to support the companies like Omega and Lallemand that are following the science and innovating.
Being a homebrewer- so far I’ve only used the Bananza and Sundew strains. Admittedly had mixed results, but that was mostly due to recipe formulation. I don’t recommend a plain hefeweizen with Bananza… haha it was boooooring. My hazy IPA with Sundew and Mosaic/El Dorado was my favorite beer I made last year though- lot’s of tropical fruit/passionfruit and some very nice pear notes too.
Hoping to get my hands on some of the IRC7 yeasts soon!
I was wondering if you used the Bananza strain in Sapwood’s The New Milkshake collab with Suspended because definitely felt that banana mouthful but doesn’t seem like you did. When will Sapwood use the Bananza strain?
Hey Scott, I brewed some Cosmic Debris – 3 gallons IPA wort with 1st wort hopping of Warrior, then citra and Simcoe steeped at 180 F post boil. Added 1 gallon Moscato grape must. Pitched Omega Cosmic Punch yeast. And voila, accidental hazy, juicy beer. Popular with friends and family. Thanks!
The one thing I’d argue is editing a fungi strain like yeast, especially transgenically seems inherently more risky than traditional GMO foods. You do have things like pathogenic yeasts so ya the Jurassic park truism is not entirely inappropriate imo. For example, Candida auris is claiming an estimated 700k lives a year, worldwide.